Selecting a model¶
This document does not describe a methodology to exactly select a drug model for a specific population. The selection in itself is a vast subject, more generic than Tucuxi implementations.
Here we focus on the kind of information required to actually build a Tucuxi drug model based on a population pharmacokinetic model published in litterature.
All PK models implemented in Tucuxi are based on compartments and are described by their differential equations (or equivalent analytical functions). These models are described in section Tucuxi PK models. If you find a very nice model you really want to have for your practice, and you discover the associated PK model is not currently supported in Tucuxi, please contact the development team. For sure there is a way to add a new PK model within the software.
So, every population PK paper is not necessarily well suited for Tucuxi. For instance some papers are only trying to describe the maximum concentration and the Area Under Curve on 24 hours, but do not offer any information about real PK parameters. Those papers shall not be used to build a model for Tucuxi.
A suitable paper shall supply the following information:
Identification of the PK model (linear or Michalis-Menten elimination for instance, type of absorption)
Covariates
Name of the covariate
Unit of the covariate
PK parameters
Name of the parameter
Mean value of the parameter
Variability of the parameter
Distribution (normal, lognormal, …)
Standard deviation, variance or coefficient of variation
A priori computation of the parameter based on the covariates
This is not mandatory, and depends on the model
Residual error model
Type of error model (Additive, proportional, mixed, …)
Sigmas associated to the error model (the number vary depending on the type)
All the numbers are usually found in a table within the paper, and the rest is to be found in the text.
Here is an example of such table, together with the a priori parameters formula, found in Fuchs2014:
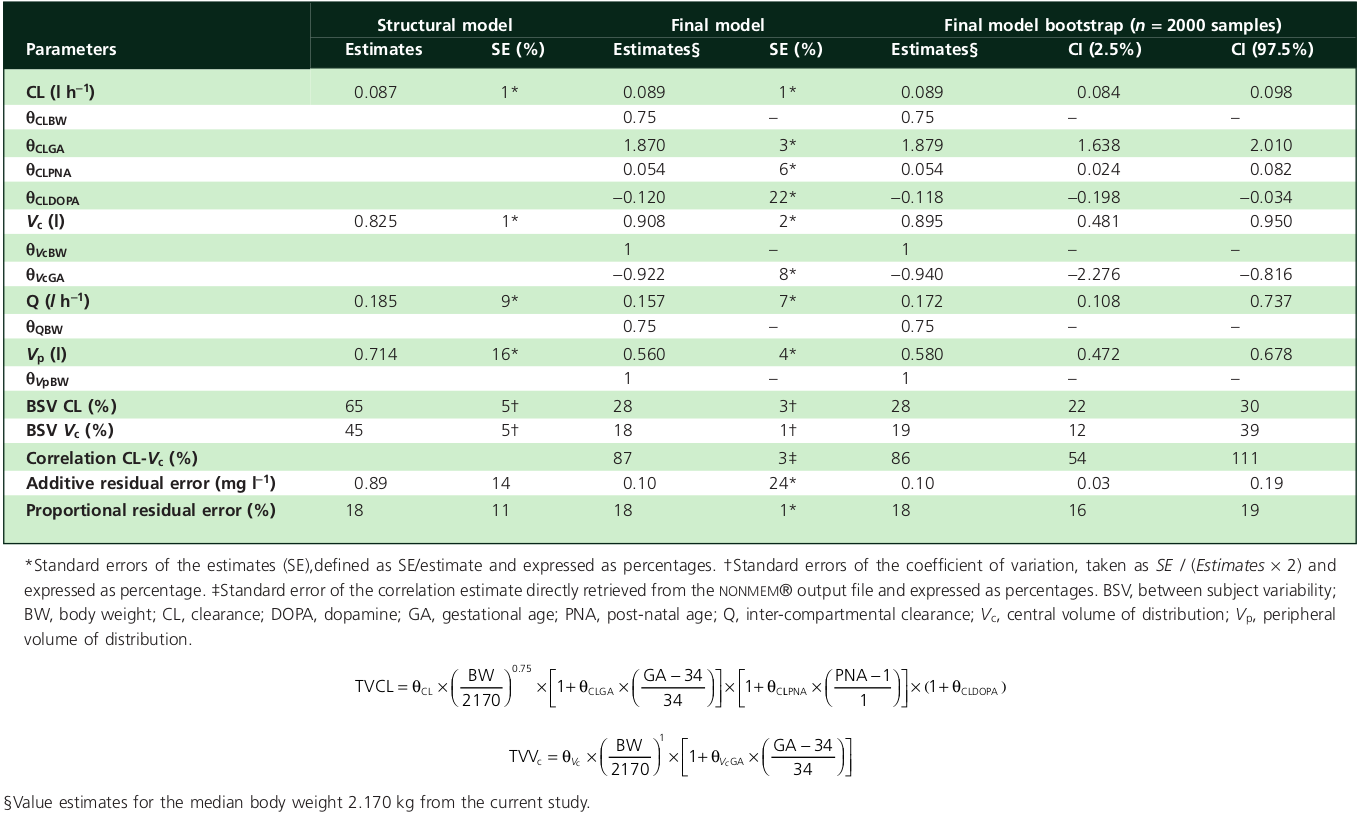
The values that shall be extracted there are the ones in the final model estimates column.
This example shows a kind of ideal representation. Depending on the paper, the presentation is not always as good as this (the world needs a standardization of population PK studies).
From all that information, you can start to use the Tucuxi drug editor to edit a new model and play with it.
Please not that the absence of variability is tolerated, but will end up with no percentiles and an inaccurate a posteriori prediction, as the Bayesian engine responsible for the calculation of PK parameters from the covariates and the measures exploits the parameters variability and the residual error model.
Finally, the drug model file shall contain information about the type of intake and available doses. This information has to be filled based on information usually not contained within a population PK paper.